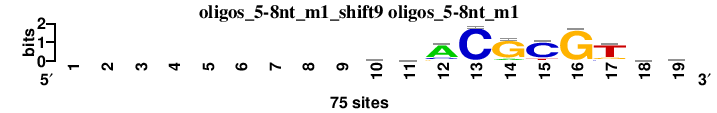
| oligos_5-8nt_m1_shift9 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=10; shift=9; ncol=19; ---------myACGCGTrk
; Alignment reference
a 0 0 0 0 0 0 0 0 0 23 18 56 0 9 5 2 0 24 17
c 0 0 0 0 0 0 0 0 0 27 21 11 73 0 60 0 8 14 10
g 0 0 0 0 0 0 0 0 0 10 13 8 0 62 0 72 11 20 27
t 0 0 0 0 0 0 0 0 0 15 23 0 2 4 10 1 56 17 21
|
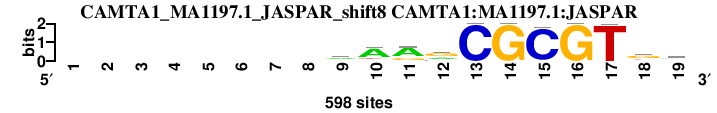
| CAMTA1_MA1197.1_JASPAR_shift8 (CAMTA1:MA1197.1:JASPAR) |
 |
0.847 |
0.706 |
5.692 |
0.920 |
0.936 |
7 |
3 |
2 |
7 |
7 |
5.200 |
2 |
; oligos_5-8nt_m1 versus CAMTA1_MA1197.1_JASPAR (CAMTA1:MA1197.1:JASPAR); m=2/25; ncol2=12; w=10; offset=-1; strand=D; shift=8; score= 5.2; --------aAarCGCGTkd
; cor=0.847; Ncor=0.706; logoDP=5.692; NsEucl=0.920; NSW=0.936; rcor=7; rNcor=3; rlogoDP=2; rNsEucl=7; rNSW=7; rank_mean=5.200; match_rank=2
a 0 0 0 0 0 0 0 0 301 430 405 172 0 0 0 0 0 63 235
c 0 0 0 0 0 0 0 0 56 46 6 140 598 0 578 0 0 58 33
g 0 0 0 0 0 0 0 0 117 45 132 285 0 598 0 598 3 326 156
t 0 0 0 0 0 0 0 0 124 77 55 1 0 0 20 0 595 151 174
|
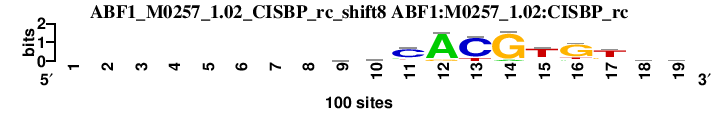
| ABF1_M0257_1.02_CISBP_rc_shift8 (ABF1:M0257_1.02:CISBP_rc) |
 |
0.758 |
0.690 |
0.247 |
0.916 |
0.929 |
8 |
4 |
25 |
8 |
8 |
10.600 |
8 |
; oligos_5-8nt_m1 versus ABF1_M0257_1.02_CISBP_rc (ABF1:M0257_1.02:CISBP_rc); m=8/25; ncol2=11; w=10; offset=-1; strand=R; shift=8; score= 10.6; --------vmCACGTGThc
; cor=0.758; Ncor=0.690; logoDP=0.247; NsEucl=0.916; NSW=0.929; rcor=8; rNcor=4; rlogoDP=25; rNsEucl=8; rNSW=8; rank_mean=10.600; match_rank=8
a 0 0 0 0 0 0 0 0 26 25 13 90 0 9 9 9 7 25 21
c 0 0 0 0 0 0 0 0 28 40 71 0 82 0 10 0 7 33 37
g 0 0 0 0 0 0 0 0 28 19 4 10 0 91 8 75 18 16 21
t 0 0 0 0 0 0 0 0 18 16 12 0 18 0 73 16 68 26 21
|
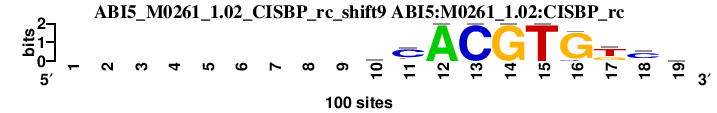
| ABI5_M0261_1.02_CISBP_rc_shift9 (ABI5:M0261_1.02:CISBP_rc) |
 |
0.715 |
0.715 |
4.663 |
0.895 |
0.890 |
17 |
1 |
9 |
13 |
14 |
10.800 |
9 |
; oligos_5-8nt_m1 versus ABI5_M0261_1.02_CISBP_rc (ABI5:M0261_1.02:CISBP_rc); m=9/25; ncol2=10; w=10; offset=0; strand=R; shift=9; score= 10.8; ---------mCACGTGkcv
; cor=0.715; Ncor=0.715; logoDP=4.663; NsEucl=0.895; NSW=0.890; rcor=17; rNcor=1; rlogoDP=9; rNsEucl=13; rNSW=14; rank_mean=10.800; match_rank=9
a 0 0 0 0 0 0 0 0 0 34 7 100 0 0 0 0 8 17 31
c 0 0 0 0 0 0 0 0 0 34 71 0 100 0 0 8 0 65 25
g 0 0 0 0 0 0 0 0 0 16 7 0 0 100 0 92 31 9 26
t 0 0 0 0 0 0 0 0 0 16 15 0 0 0 100 0 61 9 18
|
| ABI5_MA0931.1_JASPAR_rc_shift9 (ABI5:MA0931.1:JASPAR_rc) |
 |
0.714 |
0.714 |
4.671 |
0.895 |
0.890 |
18 |
2 |
8 |
14 |
15 |
11.400 |
11 |
; oligos_5-8nt_m1 versus ABI5_MA0931.1_JASPAR_rc (ABI5:MA0931.1:JASPAR_rc); m=11/25; ncol2=10; w=10; offset=0; strand=R; shift=9; score= 11.4; ---------mCACGTGkcv
; cor=0.714; Ncor=0.714; logoDP=4.671; NsEucl=0.895; NSW=0.890; rcor=18; rNcor=2; rlogoDP=8; rNsEucl=14; rNSW=15; rank_mean=11.400; match_rank=11
a 0 0 0 0 0 0 0 0 0 336 70 999 0 0 0 0 78 167 306
c 0 0 0 0 0 0 0 0 0 337 709 0 999 0 0 78 0 660 257
g 0 0 0 0 0 0 0 0 0 164 70 0 0 999 0 922 314 87 257
t 0 0 0 0 0 0 0 0 0 164 152 0 0 0 999 0 608 87 181
|




